Learning to Plot Maps with COVID19
Introduction
Well I’ve been on spring break and socially distancing in that time. It was inevitable that I would procrastinate on actual homework, which is why I took this chance to learn how to do some plotting on maps in R. Since COVID19 has become a pandemic and shut down all in person classes at my university, it’s an obvious choice for plotting I’m not going to say anything else about the virus or what people should be doing, because I am not a health care provider or epidemiologist.
Data
For this exercise, I am using the data from Johns Hopkins University Center for Systems Science and Engineering at [https://github.com/CSSEGISandData/COVID-19]. As of writing this post, the data is only updated through the 16th of March. The World Health Organization data is also available in the github repo, but I did not use it for this post.
Code
Libraries
Below are the library definitions. I tried a few different libraries for plotting maps and found the ones outline below the easiest to use.
library(tidyverse) ## Can you use R without it/.
library(RCurl) ## Used to import the data from github
library(stringr) ## Used for some minor string matching
library(lubridate) ## Used for dealing with dates
library(rnaturalearth) ## Used for mapping earth data
library(rnaturalearthdata) ## Also used for mapping earth data
library(gganimate) ## Used for animating graphsData Import and Transformation
For this project I pulled the data directly from the github repo, so I could be sure I had the most up to date data. The RCurl package makes it very simple to pull the data in through just the url. The data is stored in a wide format and all there is useful data in each file, so all three files will be pulled and then combined using dplyr.
## Retrieve all of the time series data from https://github.com/CSSEGISandData/COVID-19
## Retrieve Confirmed Cases
x <- getURL("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_19-covid-Confirmed.csv")
confirmed_cases_csse <- read.csv(text = x)
## Retrieve Confirmed Deaths
x <- getURL("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_19-covid-Deaths.csv")
confirmed_deaths_csse <- read.csv(text = x)
## Retreive Recovered Case Data
x <- getURL("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_19-covid-Recovered.csv")
confirmed_recovered_csse <- read.csv(text = x)The next step is to convert the data to a long format for easier graphing. This is really easy with the new pivot_longer function. This is the first time I have used this instead of the gather function. As someone who seemed to always have to Google how to properly do a gather, this just clicked in my brain right away. The dates were also converted using lubridate, which I have found is the easiest way to work with dates in R. As is shown in the table below the data is now in a long format that includes data from each spreadsheet.
## Convert wide to long (Praise Hadley) and convert date to lubridate
confirmed_cases_csse_gather <- confirmed_cases_csse %>%
pivot_longer(-c(Province.State, Country.Region, Lat, Long),
names_to = "Date", values_to = "Confirmed.Cases") %>%
mutate(cast_date = mdy(str_replace(Date, "X", ""))) %>% select(-Date)
## Convert wide to long (Praise Hadley) and convert date to lubridate
confirmed_deaths_csse_gather <- confirmed_deaths_csse %>%
pivot_longer(-c(Province.State, Country.Region, Lat, Long),
names_to = "Date", values_to = "Confirmed.Deaths") %>%
mutate(cast_date = mdy(str_replace(Date, "X", ""))) %>% select(-Date)
## Convert wide to long (Praise Hadley) and convert date to lubridate
confirmed_recovered_csse_gather <- confirmed_recovered_csse %>%
pivot_longer(-c(Province.State, Country.Region, Lat, Long),
names_to = "Date", values_to = "Recovered.Cases") %>%
mutate(cast_date = mdy(str_replace(Date, "X", ""))) %>% select(-Date)
## Join all data together
csse_complete <- left_join(confirmed_cases_csse_gather,
confirmed_deaths_csse_gather,
by = c('Province.State', 'Country.Region',
'Lat', 'Long', 'cast_date')) %>%
left_join(confirmed_recovered_csse_gather, by = c('Province.State', 'Country.Region',
'Lat', 'Long', 'cast_date'))
## Calculate mortality rate for each region at each day
csse_grouped <- csse_complete %>%
group_by(Country.Region) %>%
mutate(Mortality.Rate = Confirmed.Deaths / Confirmed.Cases * 100)
## Output Table
head(csse_grouped) %>% knitr::kable()| Province.State | Country.Region | Lat | Long | Confirmed.Cases | cast_date | Confirmed.Deaths | Recovered.Cases | Mortality.Rate |
|---|---|---|---|---|---|---|---|---|
| Thailand | 15 | 101 | 2 | 2020-01-22 | 0 | 0 | 0 | |
| Thailand | 15 | 101 | 3 | 2020-01-23 | 0 | 0 | 0 | |
| Thailand | 15 | 101 | 5 | 2020-01-24 | 0 | 0 | 0 | |
| Thailand | 15 | 101 | 7 | 2020-01-25 | 0 | 0 | 0 | |
| Thailand | 15 | 101 | 8 | 2020-01-26 | 0 | 2 | 0 | |
| Thailand | 15 | 101 | 8 | 2020-01-27 | 0 | 2 | 0 |
Plotting
World Map
After searching around for a bit, I found that rnaturalearth and rnaturalearthdata were the easiest packages to get world graphs generated with. As a note I created this on Linux and had some issues getting rnaturalearthhirez (which contains state level data) installed. I ended up installing the package directly from github rather than cran, which fixed my issues. Most of the settings within the ggplot I got to through trial and error. The map below shows the number of cases of COVID-19 through the size of the data points. The scale_size() function in particular was useful in tuning the size of the points on the map to what made the most sense to me. If I come back to this visualization, I would probably change the size to a proportion of the population, to more accurately show the severity of the outbreaks.
## Filter data to only the most recent day and exclude data points with 0 cases
csse_limited <- filter(csse_complete,
cast_date == ymd("2020-03-16"),
Confirmed.Cases > 0)
## Extract World data from rnaturalearth as sf
world <- ne_countries(scale = "medium", returnclass = "sf")
## Create world plot with point size related to # of cases
world_plot <- ggplot(data = world) +
geom_sf() +
geom_point(data=csse_limited, aes(x=Long,y=Lat,
size = Confirmed.Cases),
alpha = 0.25, col = "red") +
theme_bw() + theme(legend.position = "none") +
scale_size(range=c(1,10)) +
labs(x="Longitude", y = "Latitude") +
ggtitle("Confirmed COVID-19 Cases Worldwide",
"subtitle = Date:2020-03-16")
## Output plot
world_plotUnited States Map
As I live in the United States, I was interested in visualizing the current state of the confirmed cases within the United States. I’m not sure if this is the most efficient way to map this, but I simply filtered out the data to only include the united states and changed the displayed plotting area using latitude and longitude limits within the coord_sf() function.
## Filter out data points with 0 cases
csse_complete_nonzero <- filter(csse_complete, Confirmed.Cases > 0)
## Filter for data only within the United states and most recent date
csse_us <- filter(csse_complete_nonzero,
Country.Region == "US",
cast_date == ymd("2020-03-16"))
## Pull map data for United States
us_data <- ne_states(country = 'united states of america',
returnclass = "sf")
## Plot map again, but limit to just US lat and long
us_map <- ggplot(data = us_data) +
geom_sf() +
coord_sf(xlim = c(-128, -65), ylim = c(24, 50), expand = FALSE) +
geom_point(data=csse_us, aes(x=Long,y=Lat,
size = Confirmed.Cases),
alpha = 0.25, col = "red") +
theme_bw() + theme(legend.position = "none") +
labs(x="Longitude", y = "Latitude") +
ggtitle('Confirmed COVID-19 Cases United States',
subtitle = 'Date:2020-03-16') +
scale_size(range=c(2,10))
## Output map
us_mapAnimating
After graphing the data, I wanted to visualize how the case numbers had evolved over time. I looked into a couple of ways to do this, and learning Shiny and then setting up a Shiny server seemed like too much work for today, so rather than an interactive visualization, I settled on an animated plot. After searching for a few packages, gganimate seemed very straightforward, so that’s what I used.
World Map
The world map from above is recreated, however this time, I included the transition_states() function to allow the map to be animated by day. I tried to vary the speed using the transition states function by varying the transition length and state length with some mixed results. I found the simplest way to get the speed on wanted in the animation was to use the animate() function and manually set the fps (frames per second) that I wanted in the gif. There are a few discrepancies in the more recent dates, but those appear to be an issue with the data at this time and are not due to a plotting or transformation error.
Sorry if any of the animation is wonky. The code below worked well in markdown and html documents, but had some strange errors when I converted it to Jekyll for the website. So I ended up saving it as a gif and referencing that instead. This is also why a lot of the theme text sizes are changed in the graph below. (The gif generating code is not included. I just used the anim_save function.)
## Call graph again, but this time include transition_states
map <- ggplot(data = world) +
geom_sf() +
geom_point(data=csse_complete_nonzero,
aes(x=Long,y=Lat, size = Confirmed.Cases),
alpha = 0.25, col = "red") +
theme_bw() + theme(legend.position = "none") +
theme(plot.title = element_text(size=36),
plot.subtitle = element_text(size=30),
axis.title.x = element_text(size=24),
axis.title.y = element_text(size=24)) +
transition_states(cast_date,
transition_length = 2,
state_length = 1) +
labs(x="Longitude", y = "Latitude") +
ggtitle("Confirmed COVID-19 Cases Worldwide",
subtitle = "Date:{closest_state}") + scale_size(range=c(1,10))
## Display plot with manual fps
animate(map, fps = 3)
United States Map
It is then just as easy to repeat this for the United States graph. The code and graph are below.
## Filter to US only
csse_us <- filter(csse_complete_nonzero, Country.Region == "US")
## Select US data
us_data <- ne_states(country = 'united states of america', returnclass = "sf")
## Plot with added animation
us_map <- ggplot(data = us_data) +
geom_sf() +
coord_sf(xlim = c(-128, -65), ylim = c(24, 50), expand = FALSE) +
geom_point(data=csse_us, aes(x=Long,y=Lat, size = Confirmed.Cases), alpha = 0.25, col = "red") +
theme_bw() + theme(legend.position = "none") +
theme(plot.title = element_text(size=36),
plot.subtitle = element_text(size=30),
axis.title.x = element_text(size=24),
axis.title.y = element_text(size=24)) +
transition_states(cast_date,
transition_length = 2,
state_length = 1) +
labs(x="Longitude", y = "Latitude") +
ggtitle("Confirmed COVID-19 Cases United States",
subtitle = "Date:{closest_state}") +
scale_size(range=c(2,10))
## Output Plot
animate(us_map, fps = 3)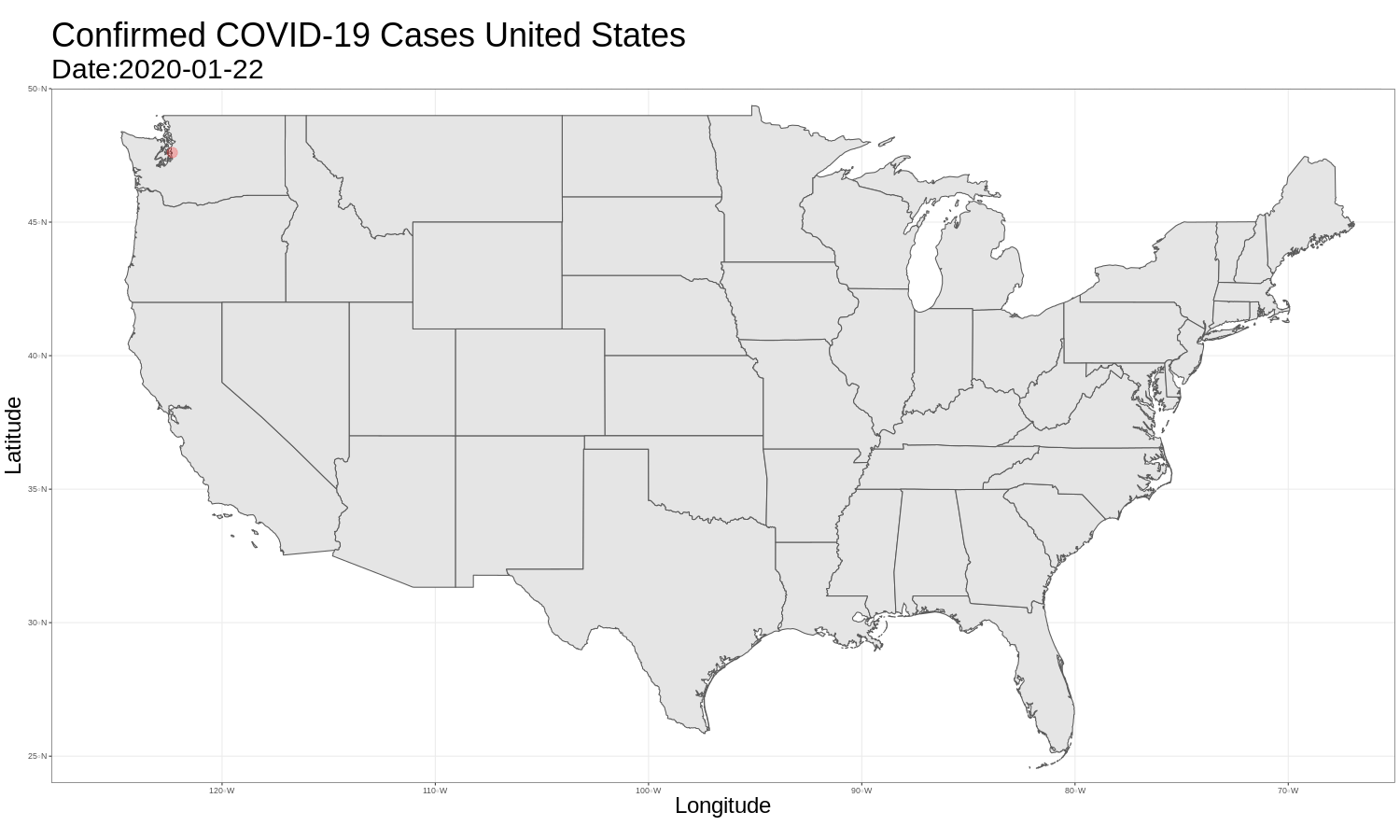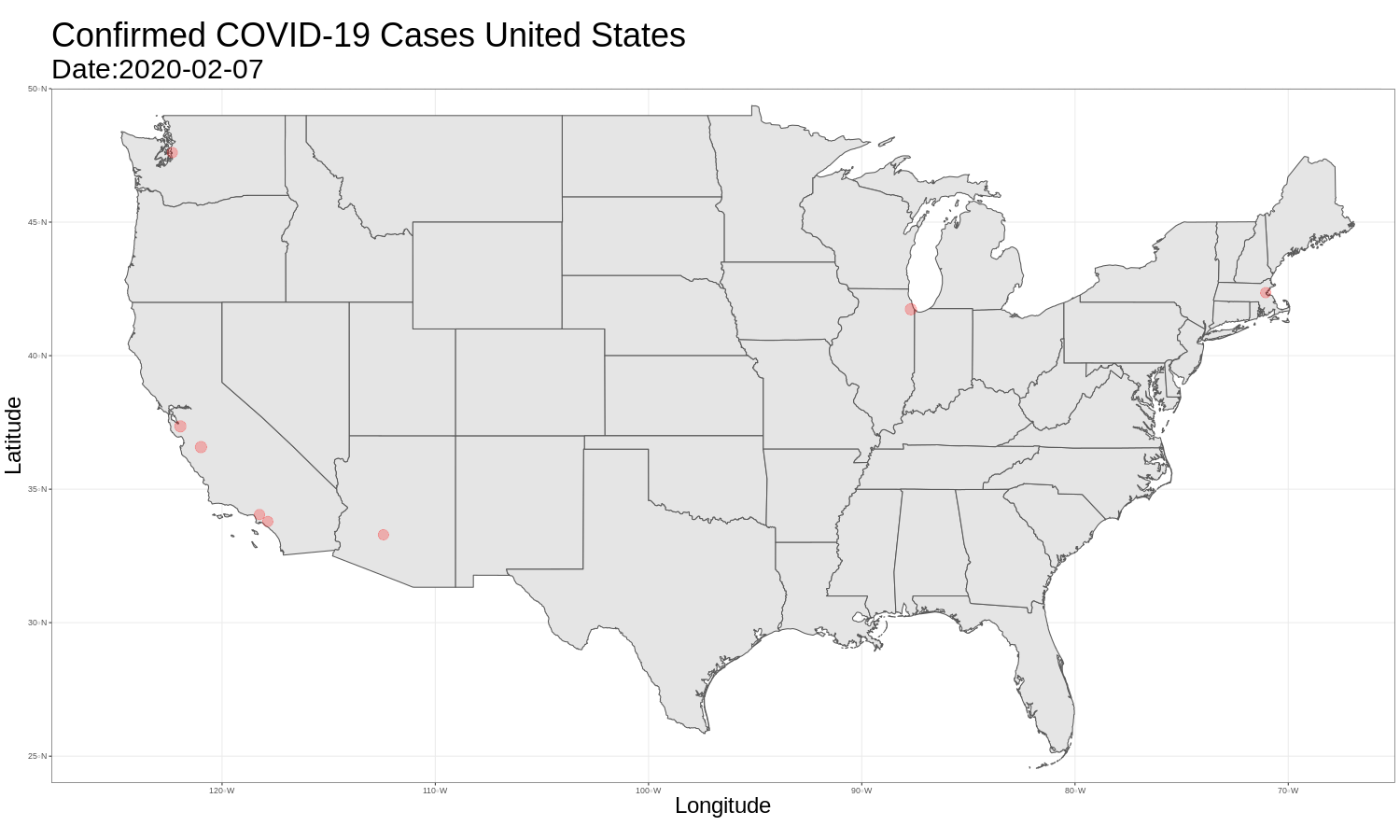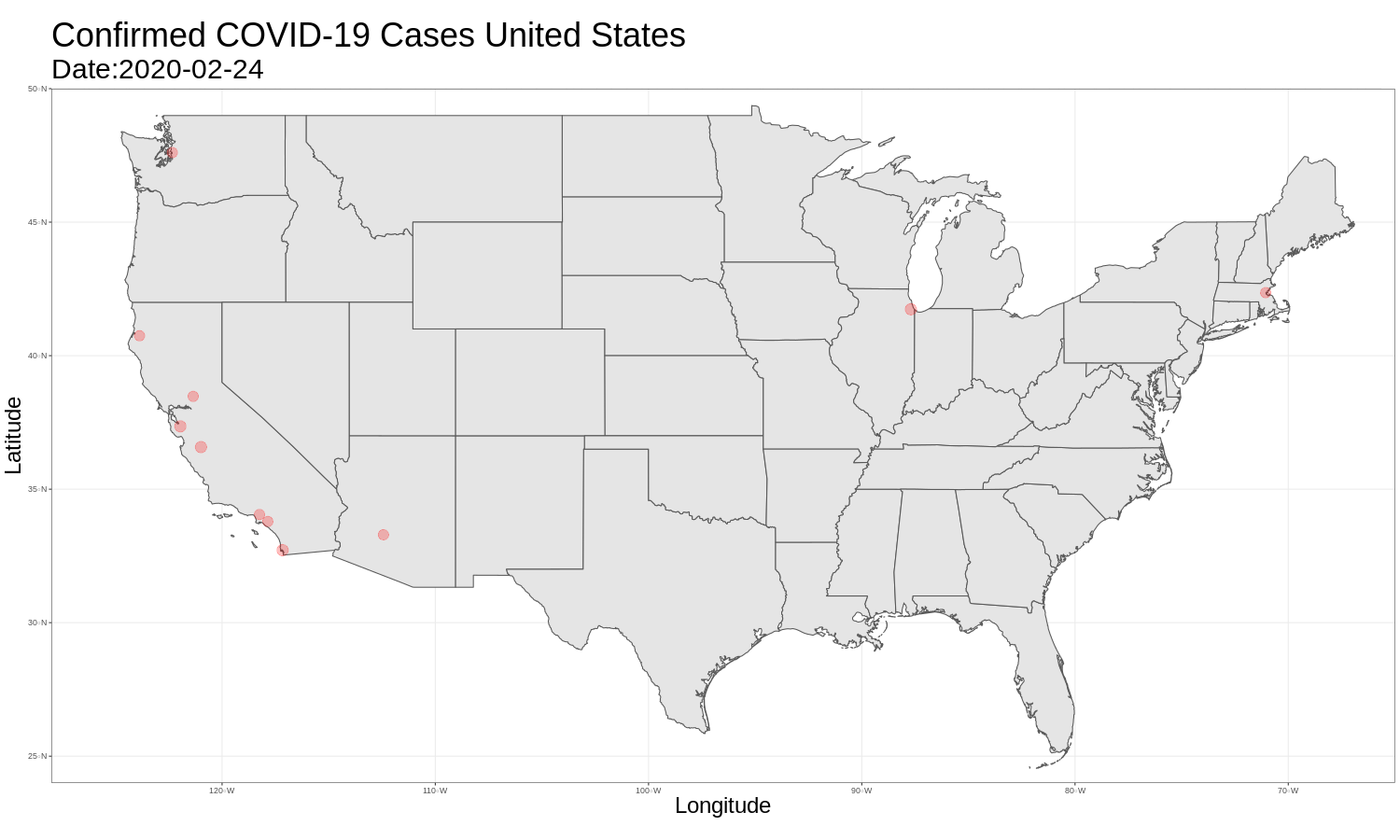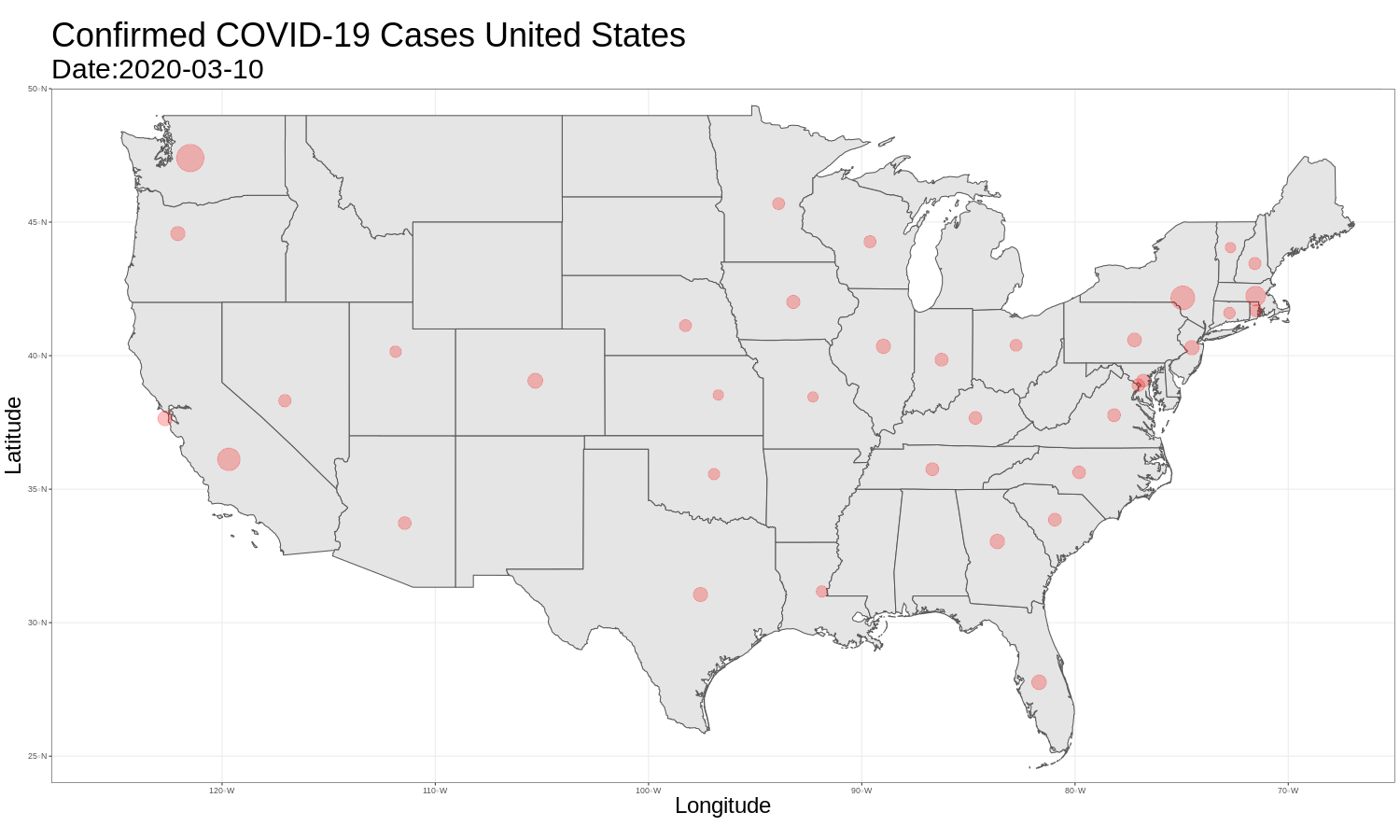
Conclusion
Overall it was actually quite easy to get started graphing maps in R. I’m looking forward to using this more as I remain socially distanced! I saw some examples that colored states based on prevalence that I would like to try out in the future. I would also potentially like to convert this to a shiny app where the timeline can be controlled with a slider. Everyone stay safe and wash your hands!


